承担的科研项目:国家自然科学基金青年基金(已结题),中国博士后基金特别资助(已结题),中国博士后基金一等资助(已结题),植物基因组学国家重点实验室青年科学家研究基金资助(已结题)。
发表论文:
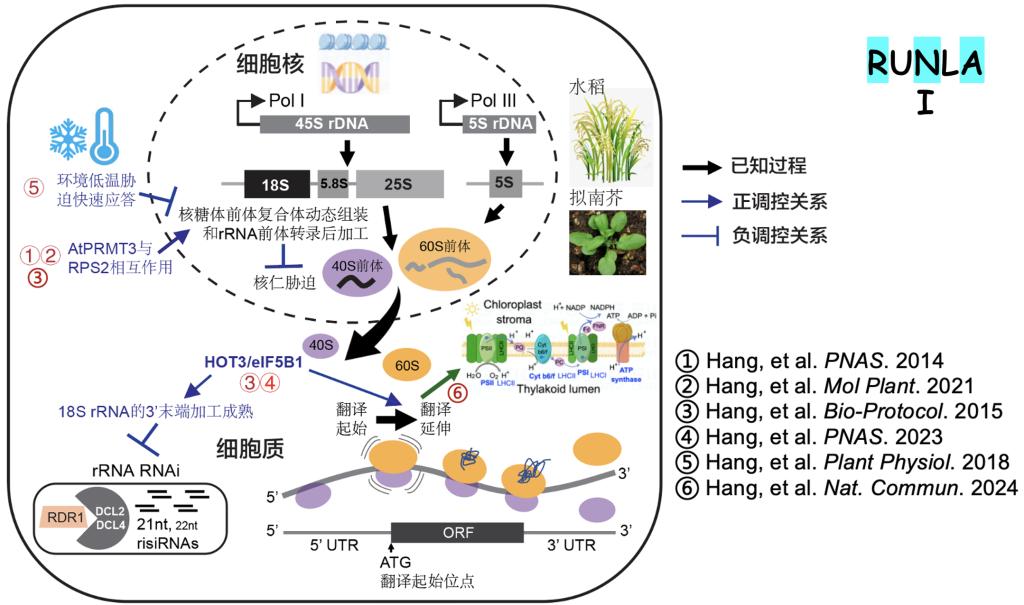
1. Hang, R*., Li, H., Liu, W., Wang, R., Hu, H., Chen, M., You, C.*, and Chen, X.* (2024). HOT3/eIF5B1 confers Kozak motif-dependent translational control of photosynthesis-associated nuclear genes for chloroplast biogenesis. Nature Communications 15, 9878. DOI: 10.1038/s41467-024-54194-1 (*共同通讯).
2. Hang, R., Xu, Y., Wang, X., Hu, H., Flynn, N., You, C., and Chen, X. (2023). Arabidopsis HOT3/eIF5B1 constrains rRNA RNAi by facilitating 18S rRNA maturation. Proc Natl Acad Sci U S A 120, e2301081120. DOI: 10.1073/pnas.2301081120.1.
3. Hang, R.#, Wang, Z.#, Yang, C., Luo, L., Mo, B., Chen, X., Sun, J., Liu, C., and Cao, X. (2021). Protein arginine methyltransferase 3 fine-tunes the assembly/disassembly of pre-ribosomes to repress nucleolar stress by interacting with RPS2B in Arabidopsis. Molecular Plant 14, 223-236. DOI: 10.1016/j.molp.2020.10.006 (#共同一作).
4. Hang, R., Liu, C., Ahmad, A., Zhang, Y., Lu, F., and Cao, X. (2014). Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc Natl Acad Sci U S A 111, 16190-16195. DOI: 10.1073/pnas.1412697111.
5. Hang, R., Wang, Z., Deng, X., Liu, C., Yan, B., Yang, C., Song, X., Mo, B., and Cao, X. (2018). Ribosomal RNA Biogenesis and its Response to Chilling Stress in Oryza sativa L. Plant Physiology 177, 381-397. DOI: 10.1104/pp.17.01714.
6. Hang, R., Deng, X., Liu, C., Mo, B. and Cao, X. (2015). Circular RT-PCR Assay Using Arabidopsis Samples. Bio-protocol 5(14): e1533. DOI: 10.21769/BioProtoc.1533.
7. You, C., He, W., Hang, R., Zhang, C., Cao, X., Guo, H., Chen, X., Cui, J., and Mo, B. (2019). FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis. Nature Communications 10, 4424. DOI: 10.1038/s41467-019-12379-z.
8. Bai, D., Zhang, J., Li, T., Hang, R., Liu, Y., Tian, Y., Huang, D., Qu, L., Cao, X., Ji, J., et al. (2016). The ATPase hCINAP regulates 18S rRNA processing and is essential for embryogenesis and tumor growth. Nature Communications 7, 12310. DOI: 10.1038/ncomms12310.
9. Wang, Z., Sun, J., Zu, X., Gong, J., Deng, H., Hang, R., Zhang, X., Liu, C., Deng, X., Luo, L., et al. (2022). Pseudouridylation of chloroplast ribosomal RNA contributes to low temperature acclimation in rice. New Phytologist 236, 1708-1720. DOI: 10.1111/nph.18479.
10. Wang, Z., Zhang, X., Liu, C., Duncan, S., Hang, R., Sun, J., Luo, L., Ding, Y., and Cao, X. (2024). AtPRMT3-RPS2B promotes ribosome biogenesis and coordinates growth and cold adaptation trade-off. Nat Communications 15, 8693. DOI: 10.1038/s41467-024-52945-8
11. Wang, X., Yu, D., Yu, J., Hu, H., Hang, R., Amador, Z., Chen, Q., Chai, J., and Chen, X. (2024). Toll/interleukin-1 receptor (TIR) domain-containing proteins have NAD-RNA decapping activity. Nat Communications 15, 2261. DOI: 10.1038/s41467-024-46499-y
12. Wang, X., Yuan, D., Liu, Y., Liang, Y., He, J., Yang, X., Hang, R., Jia, H., Mo, B., Tian, F., Chen, X., and Liu, L. (2023). INDETERMINATE1 autonomously regulates phosphate homeostasis upstream of the miR399-ZmPHO2 signaling module in maize. Plant Cell 35, 2208-2231. DOI: 10.1093/plcell/koad089.
13. Hu, H., Flynn, N., Zhang, H., You, C., Hang, R., Wang, X., Zhong, H., Chan, Z., Xia, Y., and Chen, X. (2021). SPAAC-NAD-seq, a sensitive and accurate method to profile NAD(+)-capped transcripts. Proc Natl Acad Sci U S A 118. DOI: 10.1073/pnas.2025595118.
14. Zhang, Y., Gu, L., Hou, Y., Wang, L., Deng, X., Hang, R., Chen, D., Zhang, X., Zhang, Y., Liu, C., et al. (2015). Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell Research 25, 864-876. DOI: 10.1038/cr.2015.77.
15. Yang, J., Lee, S., Hang, R., Kim, S.R., Lee, Y.S., Cao, X., Amasino, R., and An, G. (2013). OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. The Plant Journal 73, 566-578. DOI: 10.1111/tpj.12057.
![]() 蒙ICP16002391号-1
蒙ICP16002391号-1